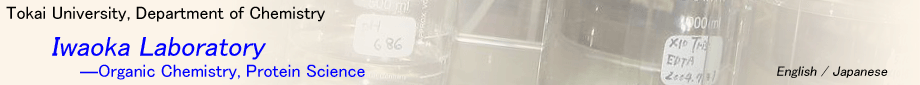Recent Activity
Grants-in-Aid for Scientific Research (KAKENHI): Scientific Research on Innovative Areas (Research under a proposed research project). (February, 2010)
How does a protein fold to the native structure? How have protein structures evolved on the earth? These questions are recognized as a folding problem in life-science fields. In our laboratory, we recently discovered that the statistical structures of amino acid residues in proteins (Ramachandran plots) follow almost perfectly the single-amino acid potentials (SAAP) in water. This coincidence means that the time-averaged structure of each amino acid in the unfolded state of a protein is almost the same to the statistically averaged structure in the native folded state. We are now working on establishing a new research paradigm based on the SAAP by pursuing the discovered unique properties of protein structures.
>Return to Activities Index
MENU
Contact Us | ©2003 Iwaoka Lab. Last Update 2015/7/28